About DBNizer
DBNizer is a tool that can be used to build discrete abstractions of dynamical systems as Dynamic Bayesian Networks (DBNs). It uses information theory measures for constructing minimal and accurate abstractions. DBNizer is written in C++ using the QT framework. The paper consists of the Apopotosis pathway case study
System overview
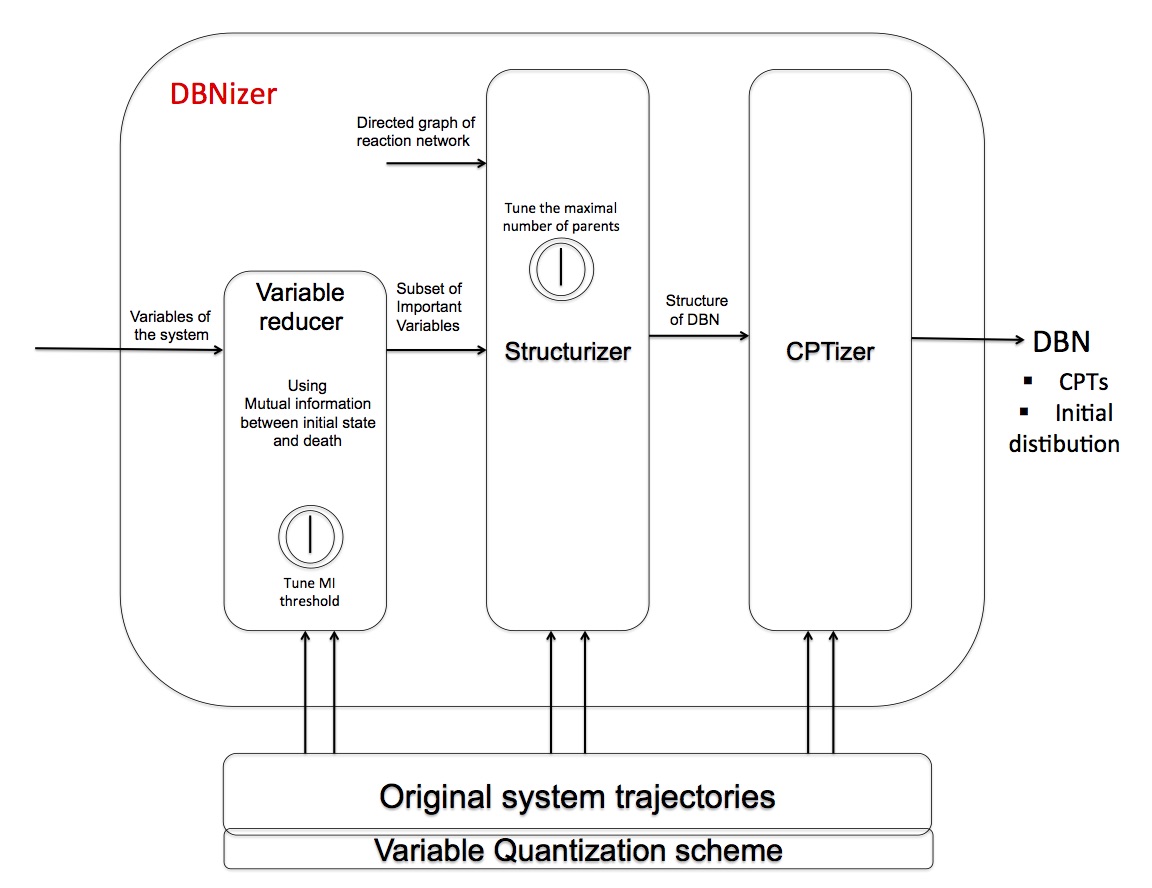
Paper reference:
Abstracting the Dynamics of Biological Pathways Using Information Theory : A Case Study of Apoptosis PathwaySucheendra K. Palaniappan, Francois Bertaux, Matthieu Pichene, Eric Fabre, Gregory Batt and Blaise Genest